Introduction to DataLad
Data Management for Open Science
August 19, 2025
About us

- PhD in neuroscience researching auditory perception
- Research software consultant at the University of Bonn
- DataLad user for ~5 years

- PhD in (cognitive) neuroscience – emotional contagion
- RDM / RSE at the Forschungszentrum Jülich
- DataLad contributor since ~4 years
Resources
- Website: olebialas.github.io/DataLad-EuroScipy25
- Contains installation instructions, slides and exercises

What is DataLad?
A community project
- 10+ years of ongoing development & maintenance 1
- 100+ contributors across core, extensions, and Handbook :
- started by:
- Michael Hanke (now: Psychoinformatics Lab, Forschungszentrum Jülich)
- Yaroslav Halchenko (now: Center for Open Neuroscience, Dartmouth College)
- owes a lot to git-annex by Joey Hess & contributors
A piece of software
- Software for data management
- Written in Python
- Based on git and git-annex
- FOSS (MIT license)
Exhaustive tracking of research components
 Well-structured datasets (using community standards), and portable computational environments — and their evolution — are the precondition for reproducibility
Well-structured datasets (using community standards), and portable computational environments — and their evolution — are the precondition for reproducibility
# turn any directory into a dataset
# with version control
% datalad create <directory># save a new state of a dataset with
# file content of any size
% datalad saveCapture computational provenance
 Which data were needed at which version, as input into which code, running with what parameterization in which computional environment, to generate an outcome?
Which data were needed at which version, as input into which code, running with what parameterization in which computional environment, to generate an outcome?
# execute any command and capture its output
# while recording all input versions too
% datalad run --input ... --output ... <command>
Exhaustive capture enables portability
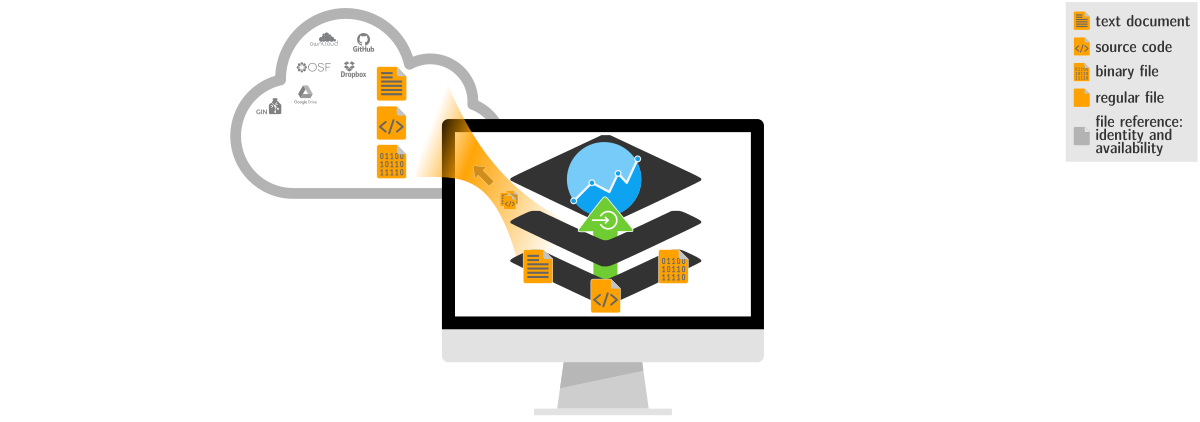 Precise identification of data and computational environments, combined for provenance records form a comprehensive and portable data structure, capturing all aspects of an investigation.
Precise identification of data and computational environments, combined for provenance records form a comprehensive and portable data structure, capturing all aspects of an investigation.
# transfer data and metadata to other sites and services with fine-grained access control for dataset components
% datalad push --to <site-or-service>Reproducibility strengthens trust
 Outcomes of computational transformations can be validated by authorized 3rd-parties. This enables audits, promotes accountability, and streamlines automated “upgrades” of outputs.
Outcomes of computational transformations can be validated by authorized 3rd-parties. This enables audits, promotes accountability, and streamlines automated “upgrades” of outputs.
# obtain dataset (initially only identity,
# availability, and provenance metadata)
% datalad clone <url># immediately actionable provenance records
# full abstraction of input data retrieval
% datalad rerun <commit|tag|range>Ultimate goal: (re)usability

Verifiable, portable, self-contained data structures that track all aspects of an investigation exhaustively can be (re)used as modular components in larger contexts — propagating their traits
# declare a dependency on another dataset and
# reuse it at particular state in a new context
% datalad clone -d <superdataset> <path-in-dataset>Hands-on: Working with a DataLad Dataset
- Open the Exercises Part 1 on the tutorial website 📖
- Each section starts with a table that contains all required commands 💻
- Feel free to chat with your neighbor and check the solutions 💡
Hands-on: Working with a DataLad Dataset

- Website: olebialas.github.io/DataLad-EuroScipy25
- Go to: Exercises > Part1: Working with DataLad Datasets